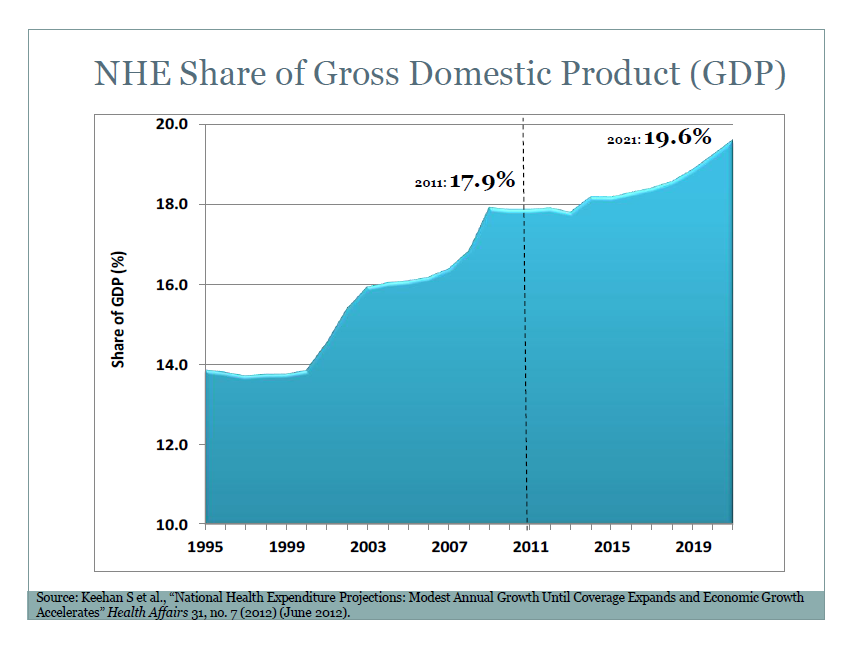
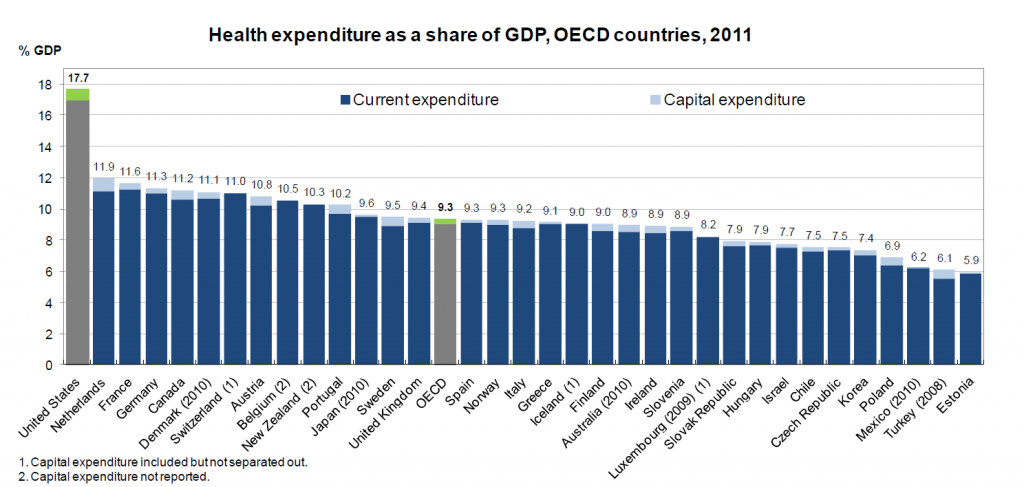
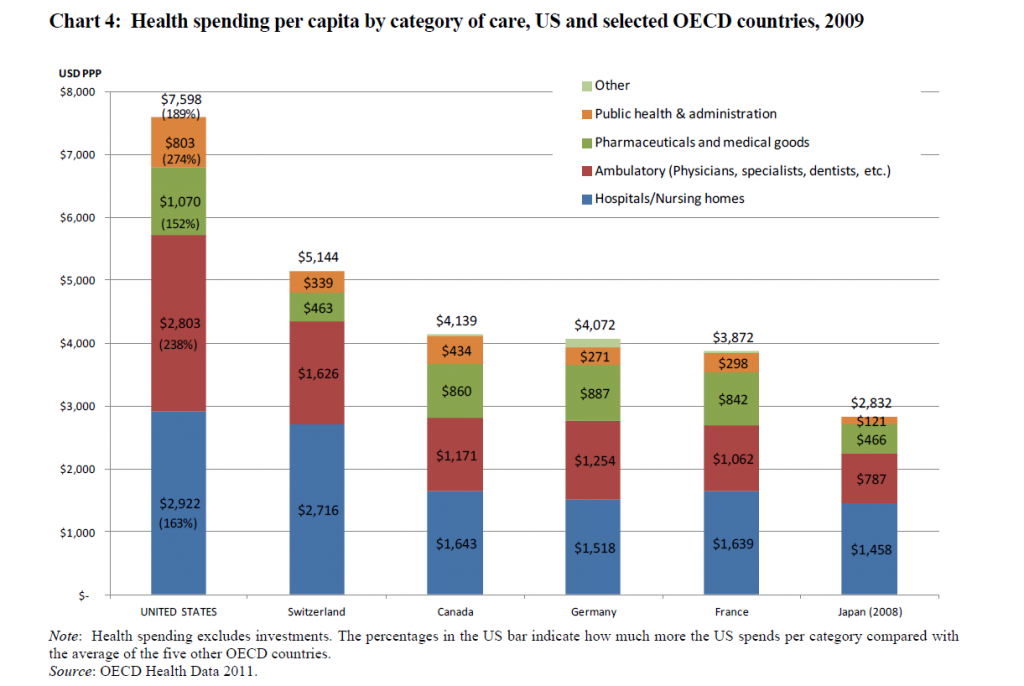
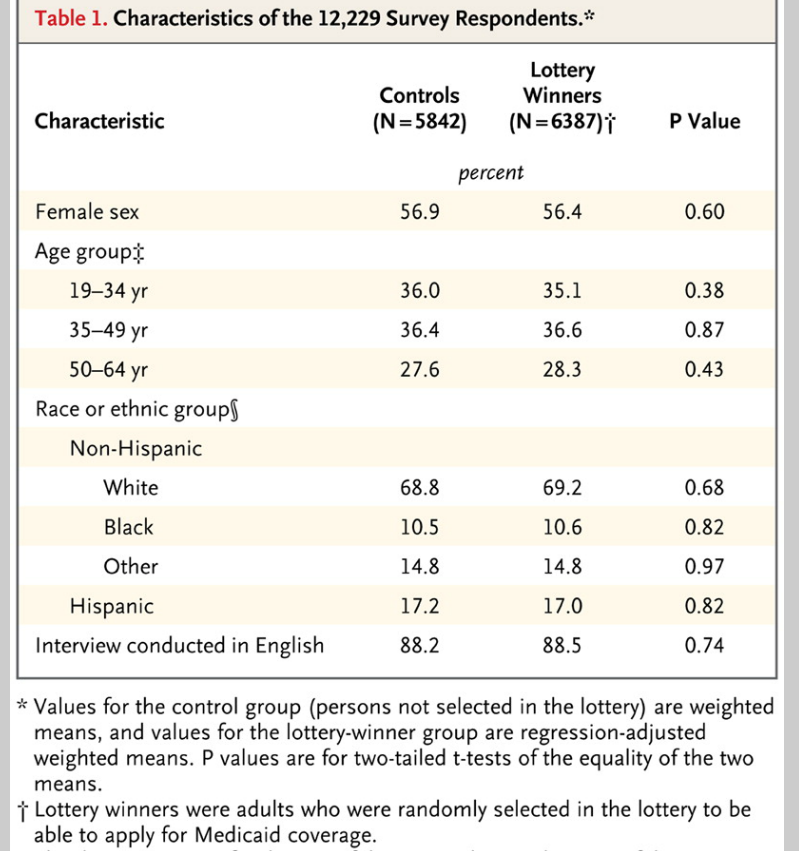
I’ve come across an interesting document – Guidance for the Use of Bayesian Statistics in Medical Device Clinical Trials developed by the Federal Drug Administration (FDA).
It’s billed as “Guidance for Industry and FDA Staff,” and provides recent (2010) evidence of the growing acceptance and success of Bayesian methods in biomedical research.
This document, which I’m just going to refer to as “the Guidance,” focuses on using Bayesian methods to incorporate evidence from prior research in clinical trials of medical equipment.
Bayesian statistics is an approach for learning from evidence as it accumulates. In clinical trials, traditional (frequentist) statistical methods may use information from previous studies only at the design stage. Then, at the data analysis stage, the information from these studies is considered as a complement to, but not part of, the formal analysis. In contrast, the Bayesian approach uses Bayes’ Theorem to formally combine prior information with current information on a quantity of interest. The Bayesian idea is to consider the prior information and the trial results as part of a continual data stream, in which inferences are being updated each time new data become available.
This Guidance focuses on medical devices and equipment, I think, because changes in technology can be incremental, and sometimes do not invalidate previous clinical trials of similar or earlier model equipment.
Thus,
When good prior information on clinical use of a device exists, the Bayesian approach may enable this information to be incorporated into the statistical analysis of a trial. In some circumstances, the prior information for a device may be a justification for a smaller-sized or shorter-duration pivotal trial.
Good prior information is often available for medical devices because of their mechanism of action and evolutionary development. The mechanism of action of medical devices is typically physical. As a result, device effects are typically local, not systemic. Local effects can sometimes be predictable from prior information on the previous generations of a device when modifications to the device are minor. Good prior information can also be available from studies of the device overseas. In a randomized controlled trial, prior information on the control can be available from historical control data.
The Guidance says that Bayesian methods are more commonly applied now because of computational advances – namely Markov Chain Monte Carlo (MCMC) sampling.
The Guidance also recommends that meetings be scheduled with the FDA for any Bayesian experimental design, where the nature of the prior information can be discussed.
An example clinical study is referenced in the Guidance – relating to a multi-frequency impedence breast scanner. This study combined clinical trials conducted in Israel with US trials,
The Guidance provides extensive links to the literature and to WinBUGS where BUGS stands for Bayesian Inference Using Gibbs Sampling.
Bayesian Hierarchical Modeling
One of the more interesting sections in the Guidance is the discussion of Bayesian hierarchical modeling. Bayesian hierarchical modeling is a methodology for combining results from multiple studies to estimate safety and effectiveness of study findings. This is definitely an analysis-dependent approach, involving adjusting results of various studies, based on similarities and differences in covariates of the study samples. In other words, if the ages of participants were quite different in one study than in another, the results of the study might be adjusted for this difference (by regression?).
An example of Bayesian hierarchical modeling is provided in approval of a device called for Cervical Interbody Fusion Instrumentation.
The BAK/Cervical (hereinafter called the BAK/C) Interbody Fusion System is indicated for use in skeletally mature patients with degenerative disc disease (DDD) of the cervical spine with accompanying radicular symptoms at one disc level.
The Summary of the FDA approval for this device documents extensive Bayesian hierarchical modeling.
Bottom LIne
Stephen Goodman from the Stanford University Medical School writes in a recent editorial,
“First they ignore you, then they laugh at you, then they fight you, then you win,” a saying reportedly misattributed to Mahatma Ghandi, might apply to the use of Bayesian statistics in medical research. The idea that Bayesian approaches might be used to “affirm” findings derived from conventional methods, and thereby be regarded as more authoritative, is a dramatic turnabout from an era not very long ago when those embracing Bayesian ideas were considered barbarians at the gate. I remember my own initiation into the Bayesian fold, reading with a mixture of astonishment and subversive pleasure one of George Diamond’s early pieces taking aim at conventional interpretations of large cardiovascular trials of the early 80’s..It is gratifying to see that the Bayesian approach, which saw negligible application in biomedical research in the 80’s and began to get traction in the 90’s, is now not just a respectable alternative to standard methods, but sometimes might be regarded as preferable.
There’s a tremendous video provided by Medscape (not easily inserted directly here) involving an interview with one of the original and influential medical Bayesians – Dr. George Diamond of UCLA.
URL: http://www.medscape.com/viewarticle/813984